GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
4.7 (430) In stock
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE

PDF) CNHplus: the chromosomal copy number heterogeneity which respects biological constraints
GitHub - DKFZ-ODCF/ACEseqWorkflow: Allele-specific copy number estimation with whole genome sequencing

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data
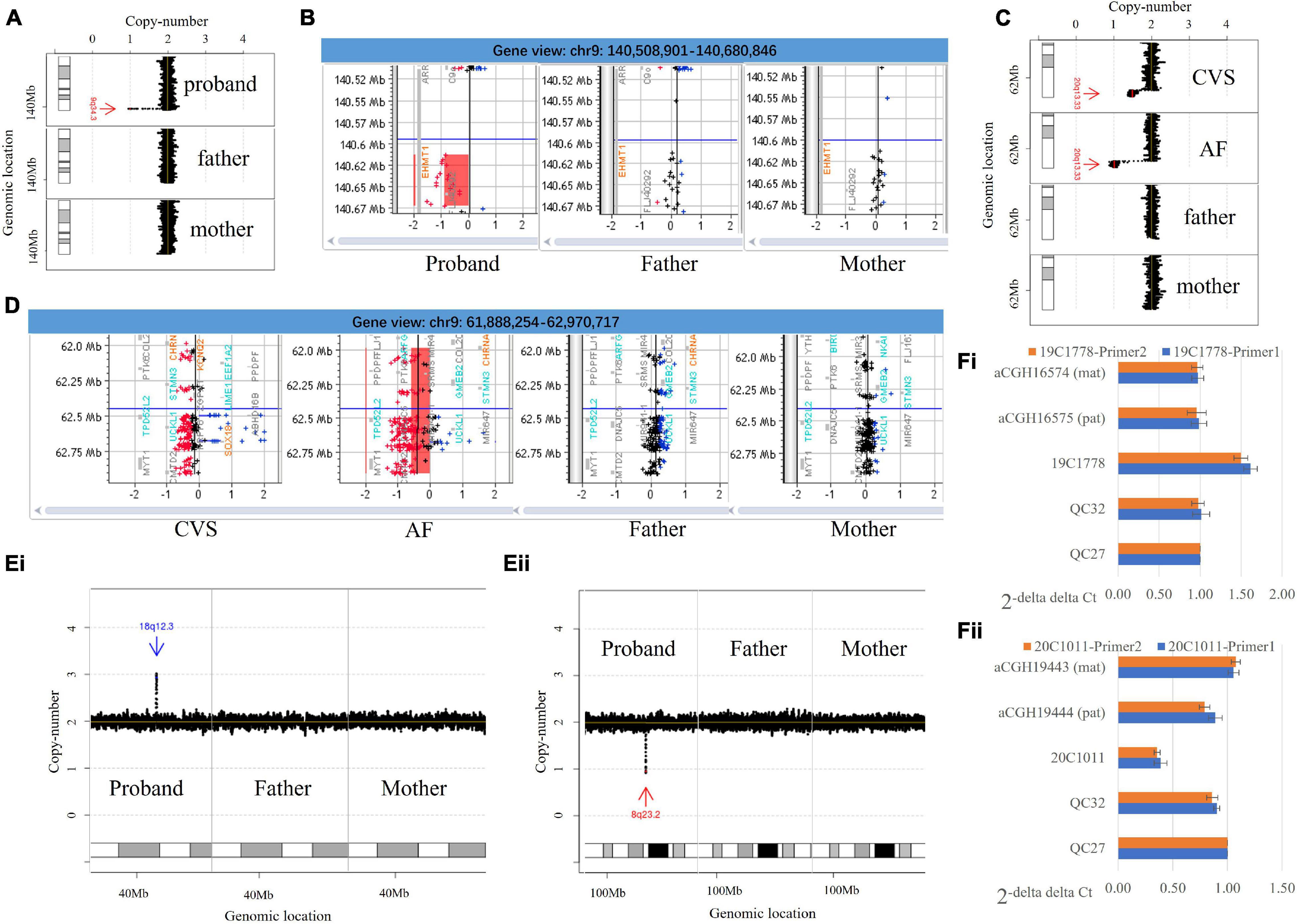
Frontiers Trio-Based Low-Pass Genome Sequencing Reveals Characteristics and Significance of Rare Copy Number Variants in Prenatal Diagnosis

PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data
GitHub - mbourgey/EBI_cancer_workshop_CNV: hands-on for NGS/SNParray CNV call trainning

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library

Copy number profiles are stable. Ultra-low pass whole genome sequencing
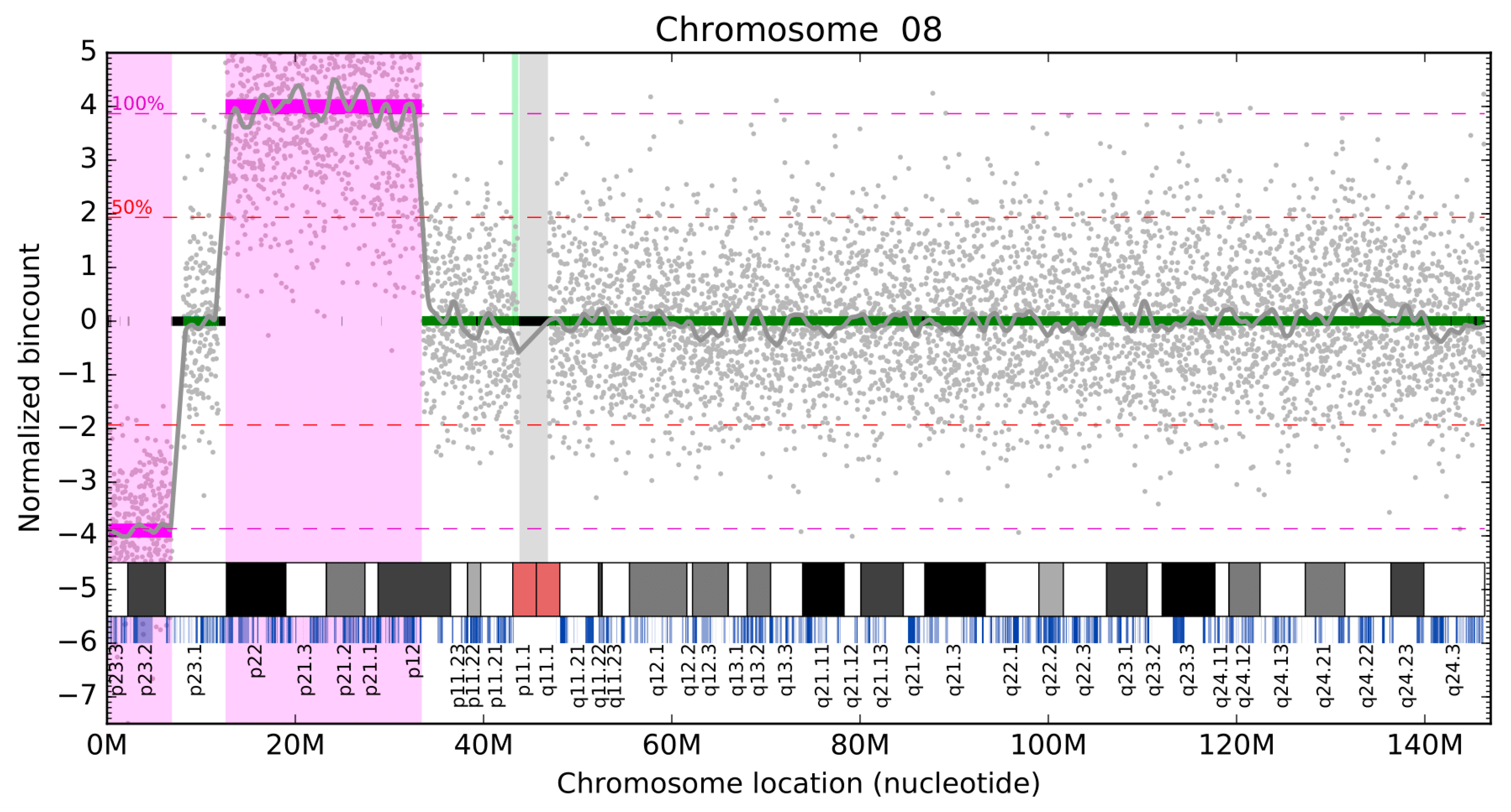
Diagnostics, Free Full-Text

Computational validation of clonal and subclonal copy number alterations from bulk tumor sequencing using CNAqc, Genome Biology

PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data
PDF) CNHplus: the chromosomal copy number heterogeneity which respects biological constraints
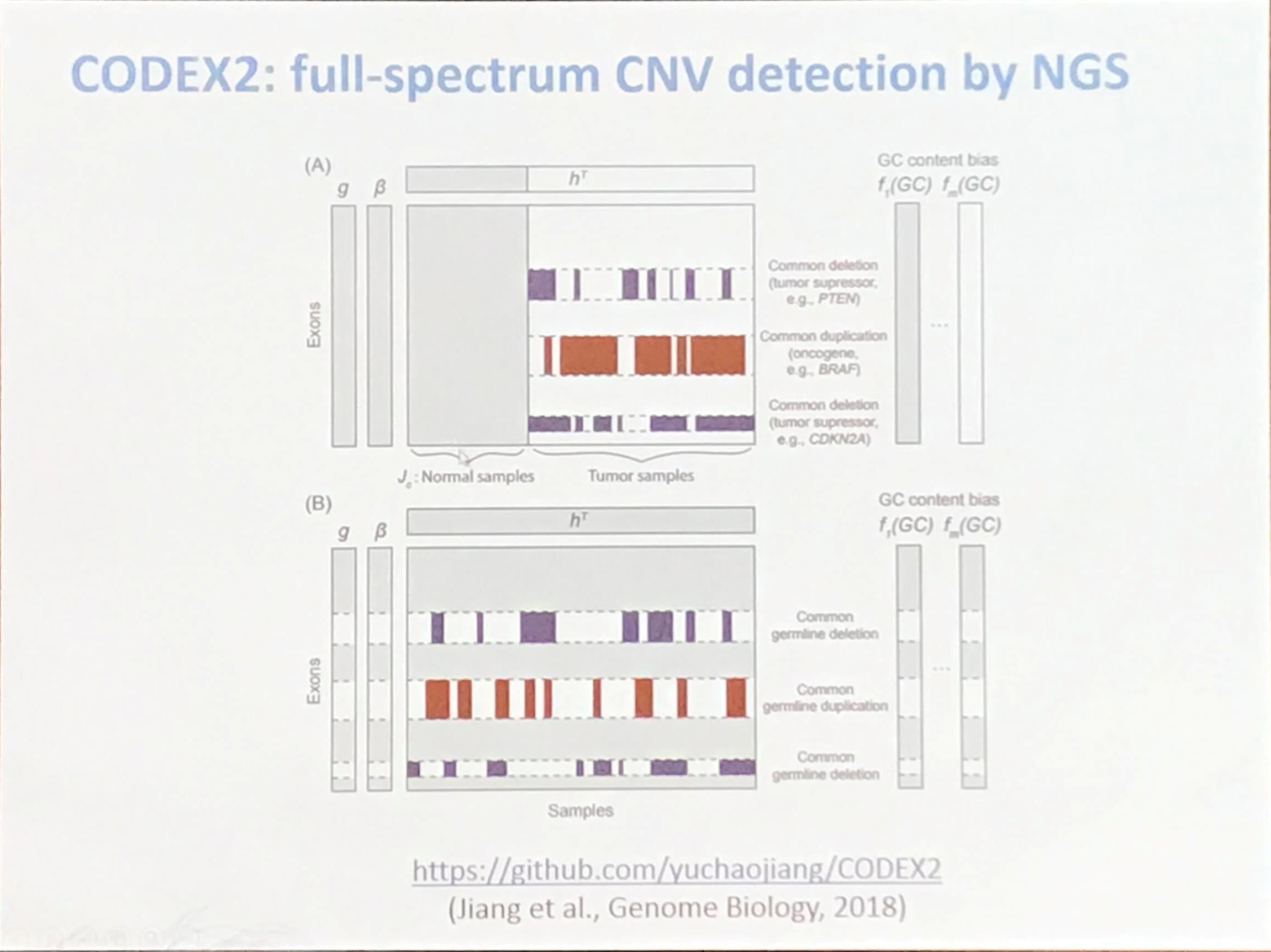
DNA copy number profiling: from bulk tissue to single cells
Understanding and increasing coverage : Knowledge Base
The variables for NGS experiments: coverage, read length, multiplexing
Extremely low-coverage sequencing and imputation increases power for genome-wide association studies
 World of Warcraft: Legion' Leveling Guide, From 0 to 110
World of Warcraft: Legion' Leveling Guide, From 0 to 110 Topper Topo De Bolo Adulto Tema Viagem Personalizado
Topper Topo De Bolo Adulto Tema Viagem Personalizado Canada Leggings-Canadian Maple Leaf Pattern Duvet Sweet Summer Casual Sling Dresses Korean Women Sexy Sundress Canada Canadian - AliExpress
Canada Leggings-Canadian Maple Leaf Pattern Duvet Sweet Summer Casual Sling Dresses Korean Women Sexy Sundress Canada Canadian - AliExpress Brandy Melville Blair Dress, Men's Fashion, Coats, Jackets and Outerwear on Carousell
Brandy Melville Blair Dress, Men's Fashion, Coats, Jackets and Outerwear on Carousell HOFISH Women Maternity Pants Plus Size Maternity Joggers Comfy Cross Waist Maternity Sweatpants Stretchy Maternity Workout Leggings Casual Loose Pregnancy Pants with Pockets, Blue,s at Women's Clothing store
HOFISH Women Maternity Pants Plus Size Maternity Joggers Comfy Cross Waist Maternity Sweatpants Stretchy Maternity Workout Leggings Casual Loose Pregnancy Pants with Pockets, Blue,s at Women's Clothing store adviicd Sport Bras for Women Full-Coverage Wirefree Bra, ComfortFlex Fit Convertible Bra for Everyday Wear Grey X-Large
adviicd Sport Bras for Women Full-Coverage Wirefree Bra, ComfortFlex Fit Convertible Bra for Everyday Wear Grey X-Large