Detecting selection in low-coverage high-throughput sequencing
4.8 (668) In stock
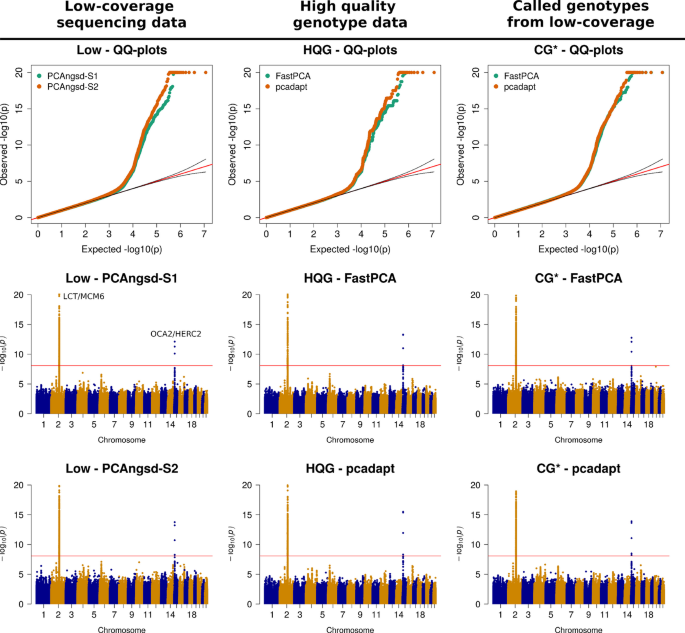
Background Identification of selection signatures between populations is often an important part of a population genetic study. Leveraging high-throughput DNA sequencing larger sample sizes of populations with similar ancestries has become increasingly common. This has led to the need of methods capable of identifying signals of selection in populations with a continuous cline of genetic differentiation. Individuals from continuous populations are inherently challenging to group into meaningful units which is why existing methods rely on principal components analysis for inference of the selection signals. These existing methods require called genotypes as input which is problematic for studies based on low-coverage sequencing data. Materials and methods We have extended two principal component analysis based selection statistics to genotype likelihood data and applied them to low-coverage sequencing data from the 1000 Genomes Project for populations with European and East Asian ancestry to detect signals of selection in samples with continuous population structure. Results Here, we present two selections statistics which we have implemented in the PCAngsd framework. These methods account for genotype uncertainty, opening for the opportunity to conduct selection scans in continuous populations from low and/or variable coverage sequencing data. To illustrate their use, we applied the methods to low-coverage sequencing data from human populations of East Asian and European ancestries and show that the implemented selection statistics can control the false positive rate and that they identify the same signatures of selection from low-coverage sequencing data as state-of-the-art software using high quality called genotypes. Conclusion We show that selection scans of low-coverage sequencing data of populations with similar ancestry perform on par with that obtained from high quality genotype data. Moreover, we demonstrate that PCAngsd outperform selection statistics obtained from called genotypes from low-coverage sequencing data without the need for ad-hoc filtering.

This table is similar to Table 2 it is the same scenario also, but
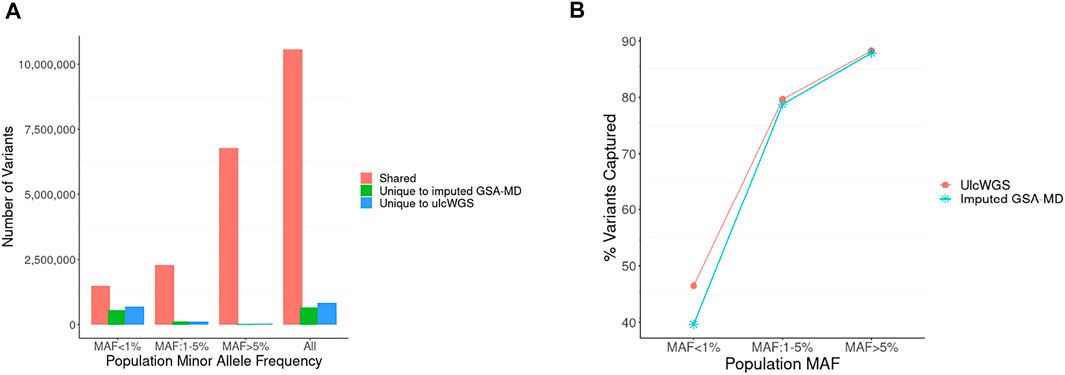
Frontiers Ultra Low-Coverage Whole-Genome Sequencing as an Alternative to Genotyping Arrays in Genome-Wide Association Studies

PDF) Detecting Selection in Low-Coverage High-Throughput Sequencing Data using Principal Component Analysis

Anders ALBRECHTSEN, Professor (Associate), PhD, University of Copenhagen, Copenhagen, Bioinformatics Centre

An Overview of Next-Generation Sequencing

Recent natural selection conferred protection against schizophrenia by non-antagonistic pleiotropy
Simulated admixture proportions for either unadmixed individuals (top)

Scatter graph showing the relationship between microsatellite length

High-throughput sequencing technologies in the detection of livestock pathogens, diagnosis, and zoonotic surveillance - ScienceDirect
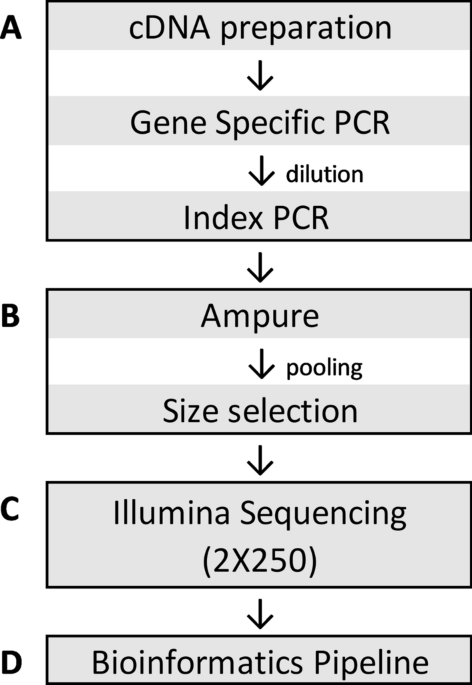
Development and validation of a high throughput SARS-CoV-2 whole genome sequencing workflow in a clinical laboratory

Rute Fonseca (@rutef) / X
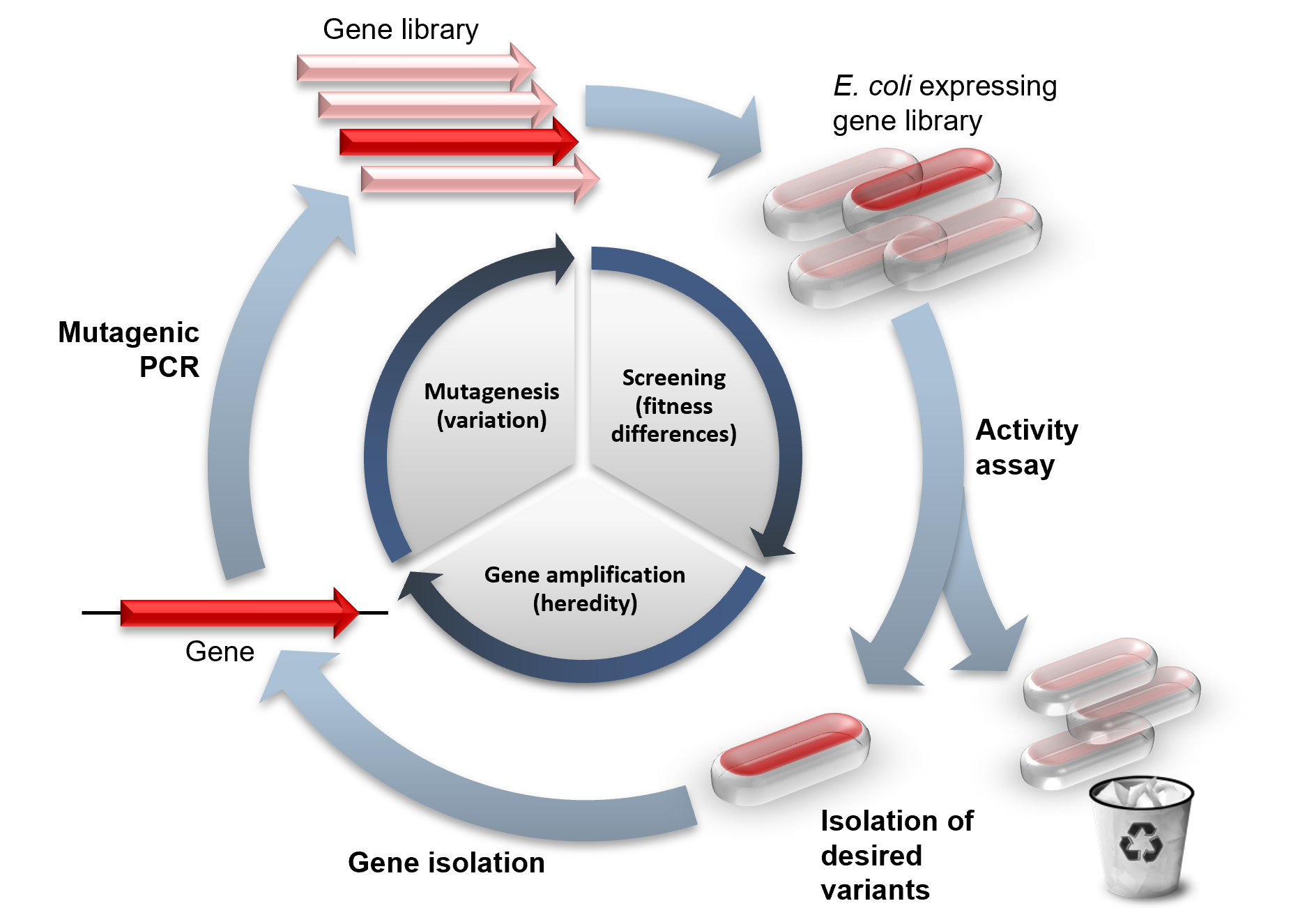
Directed evolution - Wikipedia

Anders ALBRECHTSEN, Professor (Associate), PhD, University of Copenhagen, Copenhagen, Bioinformatics Centre
Efficient phasing and imputation of low-coverage sequencing data using large reference panels
Network Booster For Low Coverage area Modelo
 Hire Formal Dresses for Your Next Event
Hire Formal Dresses for Your Next Event OVTICZA Sexy T-Back G-String Thongs for Women Plus Size Tangas Stretch Low Rise Panties Underwear XL Wine
OVTICZA Sexy T-Back G-String Thongs for Women Plus Size Tangas Stretch Low Rise Panties Underwear XL Wine- 4 Bikram Yoga Postures You Can Do Outside of the Studio — Bikram Yoga Sarasota
- What Are Brandywine Tomatoes And How Are They Best Used?
 Sexy Lace Embroidery Bodysuit Classic Hollow Push Up Bra Set Black Temptation Women Underwear Onesies Plus Size CD Cup Bodycon - AliExpress
Sexy Lace Embroidery Bodysuit Classic Hollow Push Up Bra Set Black Temptation Women Underwear Onesies Plus Size CD Cup Bodycon - AliExpress Knee pads McDavid Elite Hex para Pierna Black - Fútbol Emotion
Knee pads McDavid Elite Hex para Pierna Black - Fútbol Emotion

